I. Background
Much of my academic work has been in the field of metabolomics, the comprehensive study of small molecules in biological systems. I have also started working on leveraging Large Language Models (LLMs) to drive knowledge synthesis and automation. This interest led to my involvement in this project, alongside two undergraduate mentees, with the overarching goal of leveraging LLMs to map the landscape of metabolomics research.
The field as expanded in recent years (similar trend in many biomedical fields), driven by innovations in analytical technologies and computational methods. To manage this extensive growth, as stated above, our recent work leverages LLM, dimensionality reduction methods, and topic modeling, to visualize and interpret the sprawling landscape of metabolomics research.
Here are some of the key highlights from the preprint.
II. An Era of Accelerated Growth
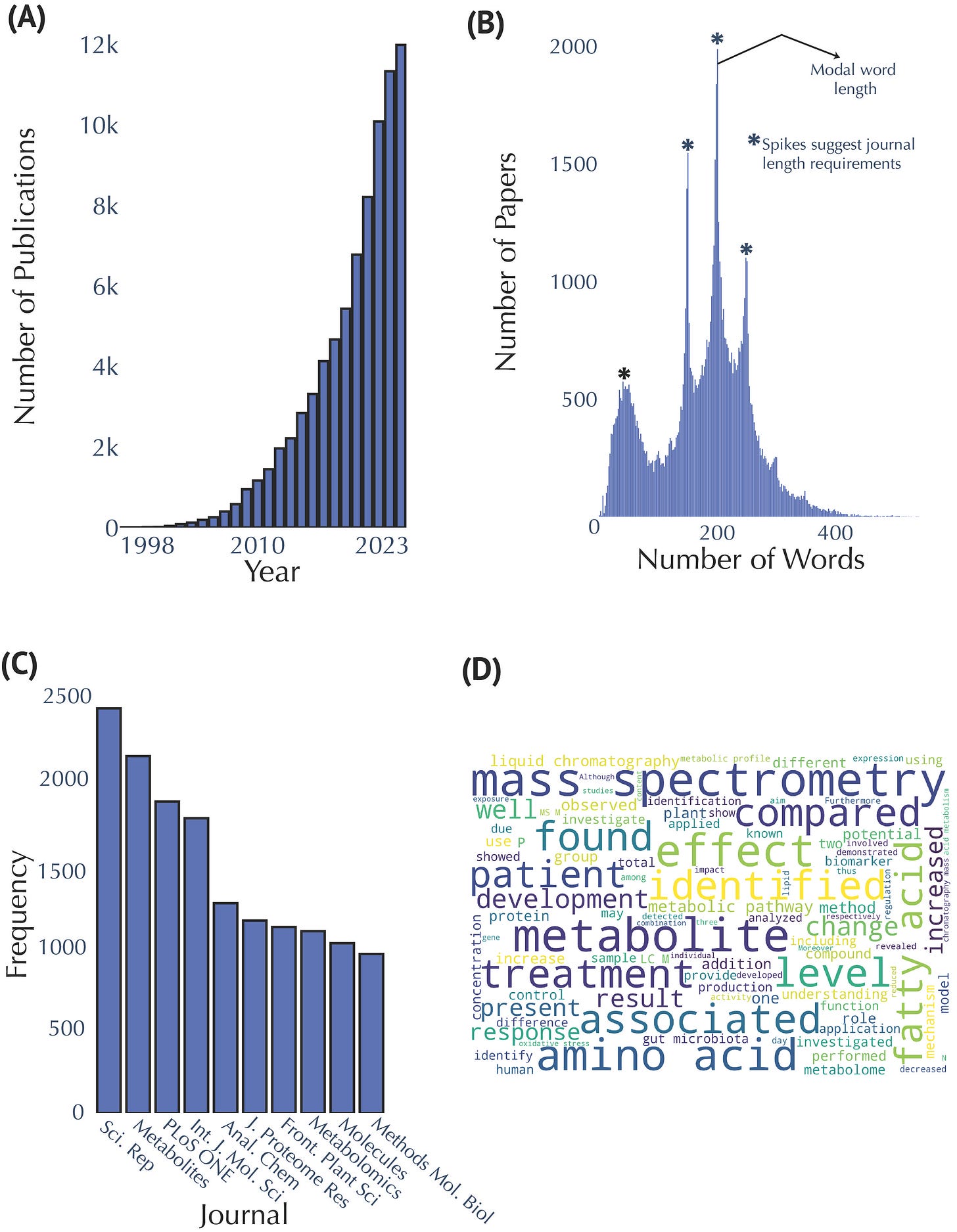
Our analysis revealed the striking growth trajectory of metabolomics. The field has had a consistent high growth rate of publication starting circa 2011, with a surge in 2020, significantly boosted by COVID-19 research.
Our data showed Scientific Reports and Metabolites as the top publishing journals, underlining the critical role of open-access platforms in disseminating metabolomics research widely. Additionally, key themes identified from abstract analyses frequently involved mass spectrometry, metabolite identification, and clinical applications, demonstrating the field's analytical and translational focus.
Publication and Abstract Analysis in Metabolomics Research (A) Annual publication counts from 1998 to 2023. (B) Distribution of abstract lengths, highlighting a modal range of 150–250 words. Spikes suggest journal word limits. (C) Top ten publishing journals in metabolomics research, with Scientific Reports and Metabolites leading the field. (D) Word cloud generated from abstracts; larger sized text indicates increased frequency of occurrence.
III. Unveiling Research Clusters through AI
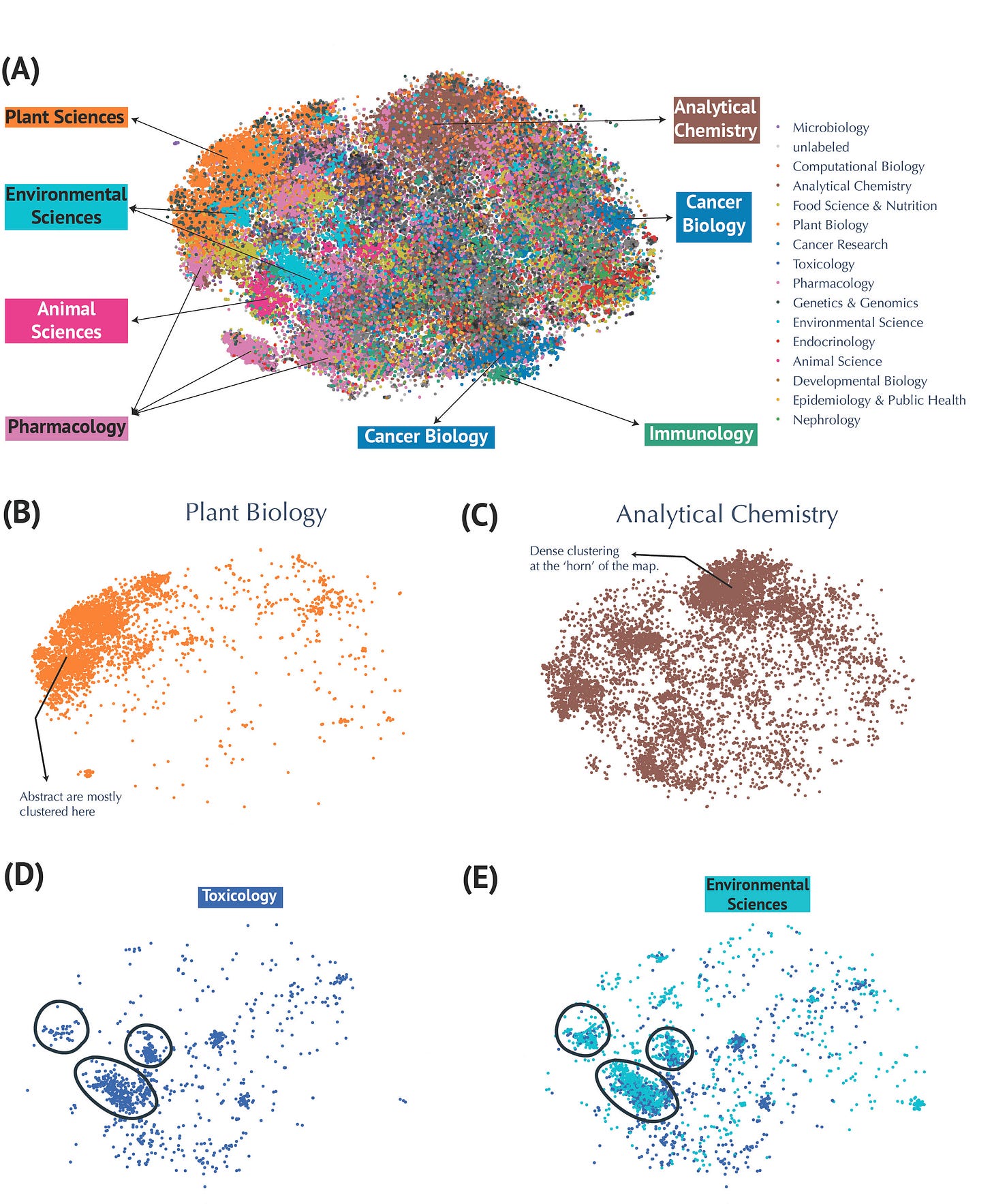
We utilized PubMedBERT—a specialized large language model trained on biomedical texts—to generate high-dimensional representations of over 80,000 abstracts. Dimensionality reduction techniques such as t-SNE provided clear visual maps revealing distinct yet interconnected research clusters (Figure 2).
Domains like plant biology formed tight, cohesive groups, whereas topics such as analytical chemistry and cancer metabolism illustrated broader, interdisciplinary connections.
Visualization of Metabolomics Research Fields using t-SNE Embeddings.
(A) Annotated t-SNE projection highlighting research domains. (B) Focused visualizations of clusters for Plant Biology and (C) Analytical Chemistry. Dense clustering is seen at the ‘horn’ of the map. (D-E) Examples of overlapping clusters, such as Environmental Sciences and Toxicology, illustrating connections on the map between related domains.
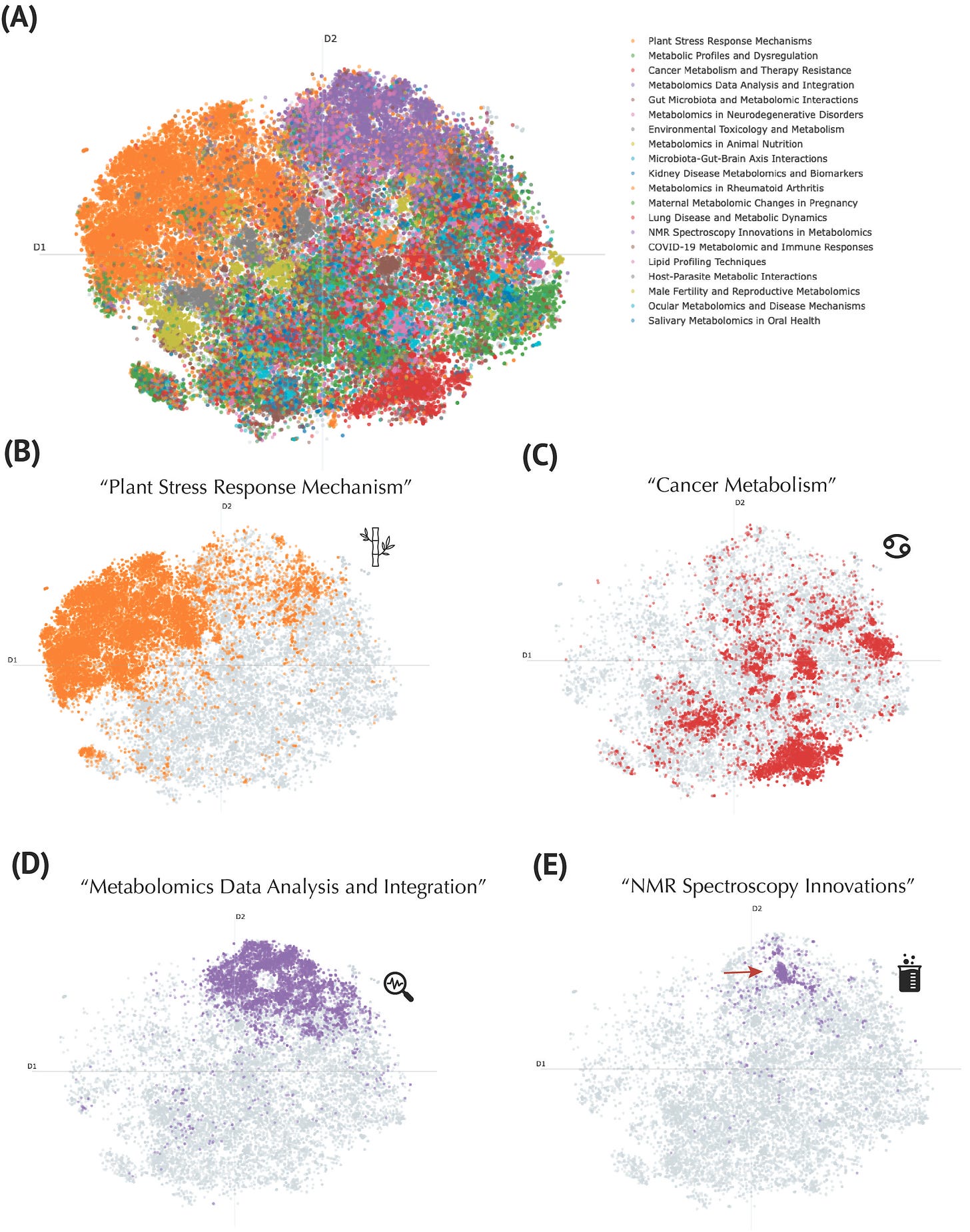
To deepen our understanding, we applied advanced topic modeling with GPT-4o mini, refining these clusters into 20 meaningful categories. This analysis went beyond traditional journal-based classifications, providing thematic labels such as "Plant Stress Response Mechanisms," "Metabolomics Data Analysis and Integration," and "NMR Spectroscopy Innovations," enhancing interpretability and utility.
Topic Modeling of the Metabolomics Corpus Using a Large Language Model. (A) Global t-SNE projection of publications, colored by representative topics discovered through the BERTopic pipeline. The legend (top right) lists exemplar thematic labels generated by GPT4o mini. (B) Focus on publications labeled as “Plant Stress Response Mechanism” (orange). (C) Cluster of “Cancer Metabolism” publications (red), illustrating the concentration of work on metabolic pathways implicated in oncogenesis and therapy resistance. (D) “Metabolomics Data Analysis and Integration” topic (purple), marking computationally oriented publications. (E) “NMR Spectroscopy Innovations” topic (purple), distinct from the data-integration cluster, focused on methodological advancements in nuclear magnetic resonance techniques.
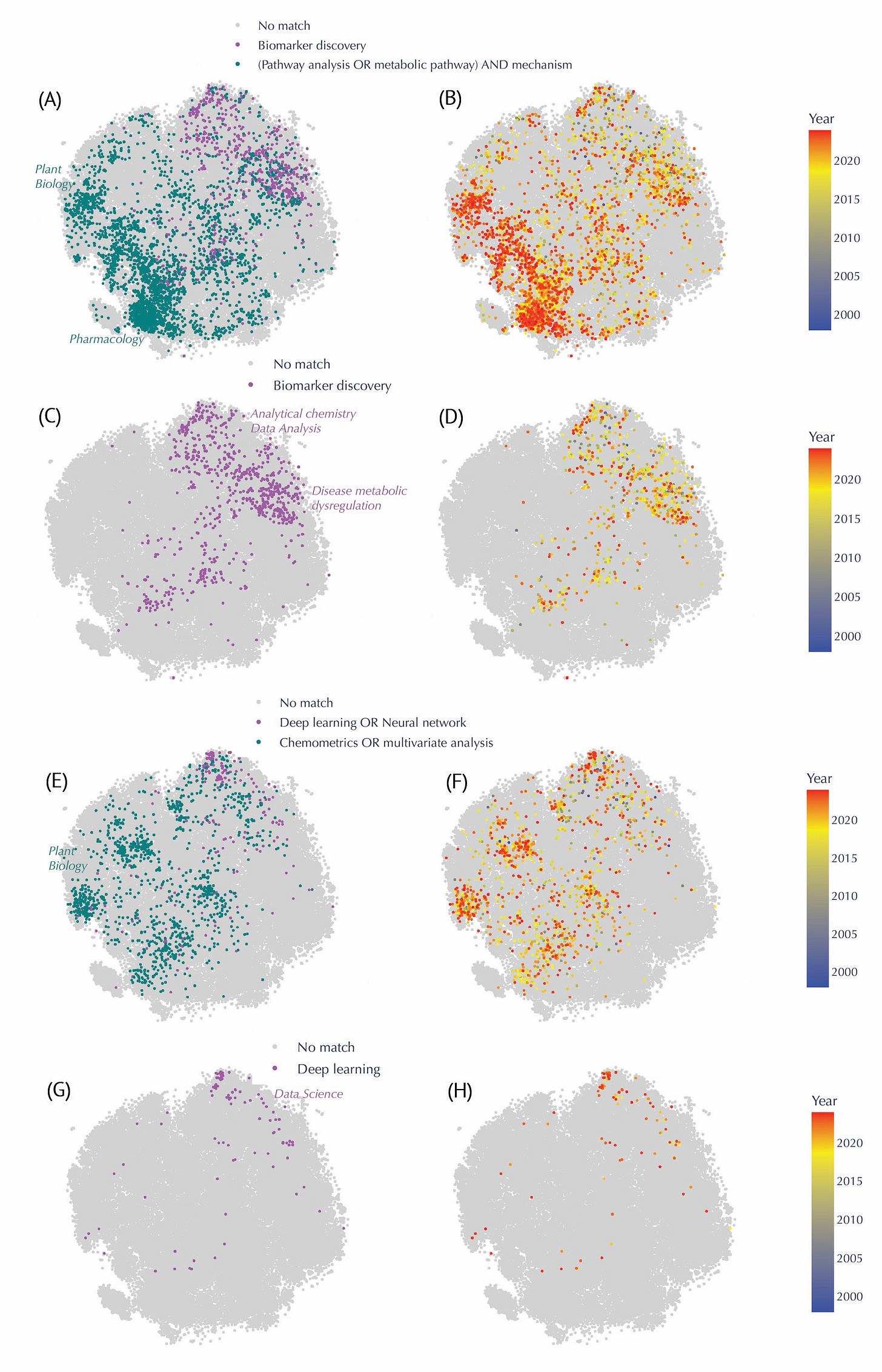
Keyword analysis within the literature further illuminated trends. "Biomarker discovery" papers spanned broad regions, particularly prominent in clinical research from the mid-2000s. In contrast, research focused on elucidating metabolic pathway mechanisms showed a concentrated presence in pharmacology and plant biology. The adoption of deep learning techniques, a recent methodological shift post-2015, also stood out clearly, reflecting contemporary technological advancements.
Evolution of Keyword-based Queries in the Metabolomics Corpus, Highlighting Shifts in Methodological Approaches and Conceptual Focuses Over Time. In each keyword panel, matched abstracts appear in color, while unmatched entries appear in light gray. The heatmap on the right panels transitions from blue (older publications) to red (more recent publications). (A–B) Comparison between “Biomarker discovery” (teal) and “(Pathway analysis OR metabolic pathway) AND mechanism” (purple). Papers mentioning “pathway mechanisms” cluster prominently in plant biology and pharmacology regions and are more recent (B), while “biomarker discovery” studies stretch across analytical chemistry and disease‐related areas from the mid‐2000s onward (A). (C–D) Focus on “Biomarker discovery” alone (purple) and its temporal distribution, revealing a surge in clinical and disease‐oriented research in the late 2000s (D). (E–F) Contrast between “Deep learning OR neural network” (purple) and “Chemometrics OR multivariate analysis” (teal). Classical multivariate methods span earlier periods, whereas deep learning publications have intensified mainly post‐2015 (F). (G–H) Spotlight on “Deep learning” (purple), showing relatively sparse coverage overall, yet relatively denser clustering in data science–focused areas in more recent years (H).
IV. Responding to Global Challenges
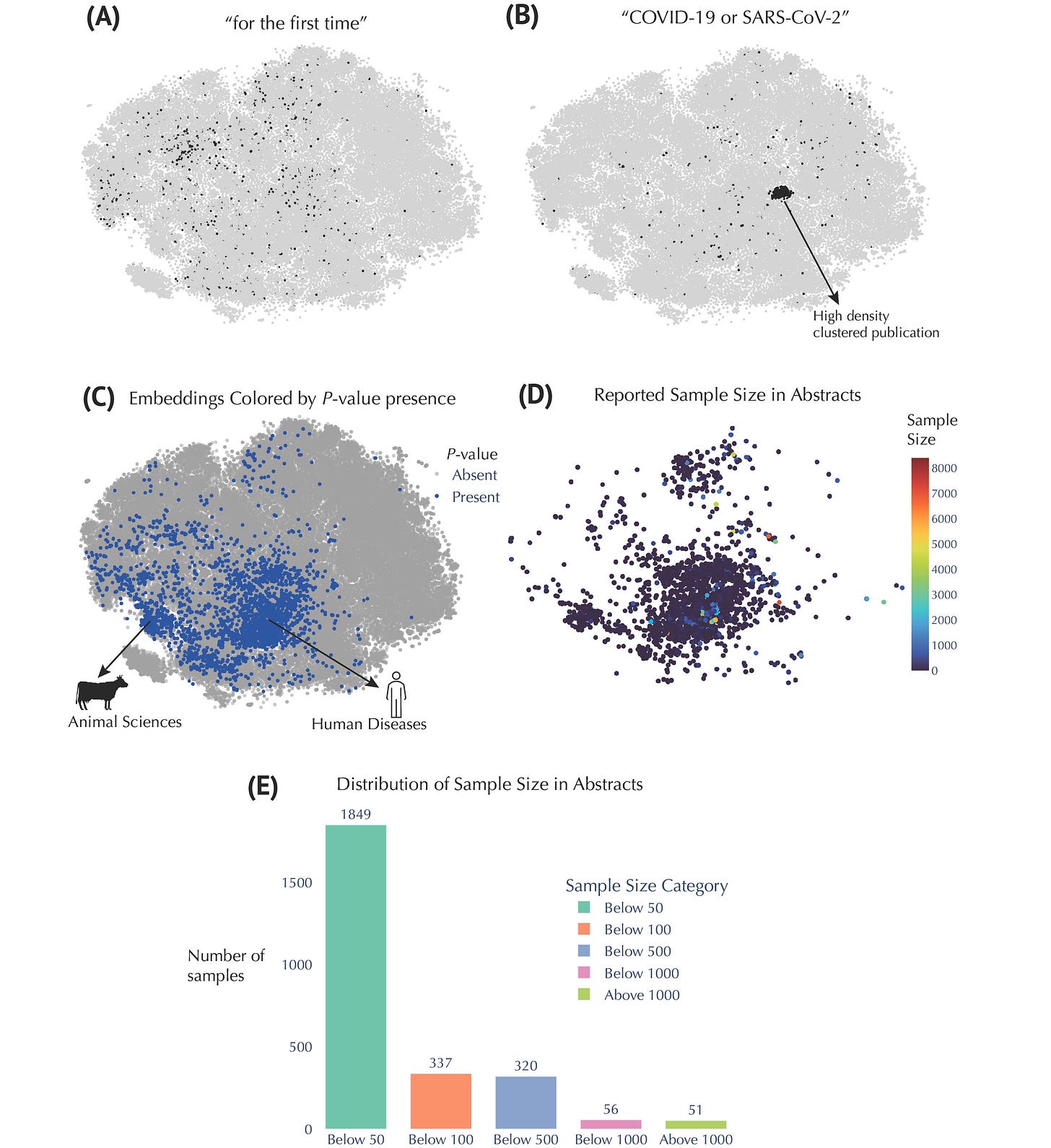
The COVID-19 pandemic profoundly impacted the metabolomics field, as illustrated by the focused cluster of research addressing pandemic-driven questions. These studies highlighted the rapid adaptability of metabolomics research in response to pressing global health challenges. Nevertheless, a persistent limitation emerged from our analysis: small sample sizes dominate the literature, suggesting an urgent need for larger, more robust clinical studies.
Insights from Embedding Metadata in Metabolomics Research. (A) t-SNE embeddings filtered to show abstracts containing the phrase “for the first time.” (B) t-SNE embeddings filtered for abstracts containing “COVID‑19” or “SARS‑CoV‑2”. (C) Embeddings colored by presence or absence of P‑values. For example, phrases such as “p<0.05,” “P=0.01,” or “p > 0.001” match this pattern. (D) Embeddings colored by reported sample size. (E) Distribution of these extracted sample sizes across the relevant abstracts.
To democratize access to these insights, we've developed an interactive online app, available at metascape.streamlit.app. Researchers can dynamically explore trends, filter literature by specific interests, and identify emerging research topics effortlessly.
In conclusion, this work demonstrates how combining natural language processing, and topic modeling can significantly enhance our understanding of scientific fields. Such an approach not only captures the evolution and diversity of research themes but also provides practical insights, helping to guide future interdisciplinary research efforts.
Here is the Preprint
Explore the metabolomics map further at our interactive web application. Watch demo here.